|
This page discusses:
Introduction Naming Intein Motifs Criteria for Intein Designation Intein Regions and Domains Intein Motif Consensus Sequences
Introduction
What is an Intein?
A large in-frame insertion in a sequenced gene that is absent in other sequenced homologs suggests that this gene may
contain an
intein. Inteins are defined by conserved motifs specific to the intein splicing domain and by the protein splicing reaction
that they perform. Inteins are single turnover enzymes and the flanking extein residues in the precursor are their 'substrate'.
Inteins are often found by running any of the commonly available sequence comparison programs
such as Bestfit, Gap or Blast. Significant Blast matches are often found to the extein protein AND one or more proteins containing
similar inteins. More sophisticated searches can be performed using intein motifs
(Pietrokovski 1994, Perler 1997 and
Pietrokovski 1998) or a Hidden Markov Model
(Dalgaard 1997 and Gorbalenya 1998).
Please note: Finding DOD homing endonuclease motifs is not sufficient evidence that the gene contains an intein since DOD homing endonucleases are also present in introns and as free standing genes.
The InBase on-line Blast server is a convenient way of confirming that a sequence contains an intein. It often yields more significant probability values than those obtained when searching larger databases, because inteins sequences are not very similar, even within their most conserved motifs.
What is Protein Splicing?
Protein splicing is defined as the excision of an intervening sequence (the INTEIN) from a protein precursor and the concomitant ligation of the flanking protein fragments (the EXTEINS) to form a mature host protein (extein) and the free intein (Perler 1994). Intein-mediated protein splicing results in a native peptide bond between the ligated exteins (Cooper 1993). Extein ligation differentiates protein splicing from other forms of autoproteolysis and conserved intein motifs differentiate inteins from other types of in-frame sequences present in one homolog and absent in another homolog.
The term 'Protein Splicing' has been associated with inteins since 1994
(Perler 1994). Recent papers have described protein rearrangements that are not intein-mediated. The mechanism of these rearrangements is currently unknown, but preliminary evidence suggests that they are mediated by various cellular enzymes. For clarity, we suggest calling these non-intein mediated events either protein rearrangements or Protein Editing. Inteins have sometimes been called 'Protein Introns'. Introns are intervening sequences that are spliced out of RNA before the mRNA is translated into a protein. The intron and the exon usually do not form a single open reading frame (ORF). During intein-mediated protein splicing, the intervening sequence is both present in the mature mRNA and translated to form a precursor protein. The intein is then spliced out of the precursor protein. The intein plus the first C-extein residue (called the +1 amino acid) contain sufficient information to mediate splicing of the intein out of the host protein and ligation of the exteins to form the active host protein. Many inteins can splice in heterologous foreign proteins if they are placed in a compatible host protein environment. The rules for what constitutes a good foreign extein environment are not well understood. It also appears that different inteins are more robust than others, with some inteins requiring many native extein residues and others only a single native C-extein amino acid.
Intein Alleles and the Phylogenetic Distribution of Inteins
The phylogenetic distribution of inteins is sporadic. The presence of an intein in a particular gene does not necessarily mean that an extein homolog from a closely related species or strain will have the same intein. For example, look at the inteins present in the 3 insertion sites in DNA polymerases from various strains of archaea or GyrA inteins in various species of Mycobacterium. At this time, it is not clear whether this pattern of intein distribution represents loss of ancient inteins or more recent acquisition of inteins due to intein gene mobility. In many cases analyzed, the codon usage and GC content of the intein coding region is different from the surrounding extein coding region, suggesting recent horizontal transmission. Organisms that have a large number of inteins may have acquired this large number of inteins because they (1) can easily take up DNA from the environment (naturally competent), (2) share viruses, conjugative elements, plasmids, etc. that have broad host ranges, or (3) have very efficient gene conversion machinery, double-stand breaks repair systems and/or recombination systems.
Return to Top
Naming Intein Motifs
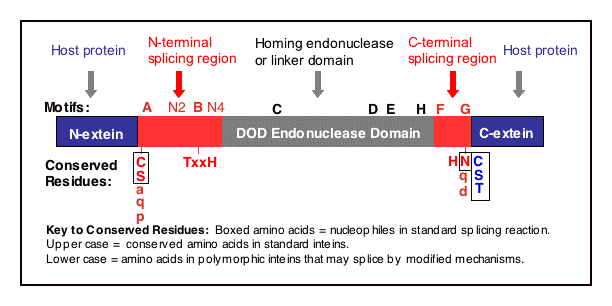
Ten intein motifs have been identified (see below for their consensus sequence): Blocks A-H (Pietrokovski 1994 and Perler 1997) and Blocks N2 and N4 (Pietrokovski 1998). Intein Blocks A, N2, B, N4, F, and G are involved in protein splicing. Intein Blocks C, D, E, and H are part of the DOD homing endonuclease domain present in many inteins. Pietrokovski 1998 suggests renaming intein Blocks as follows: A=N1, B=N3, C=EN1, D=EN2, E=EN3, H=EN4, F=C2 and G=C1. Note that hedgehog protein autoprocessing domains have conserved motifs similar to intein Blocks A and B (Koonin 1995 and Hall 1997)
Return to Top
Four Criteria for intein designation:
A combination of criteria have been used to identify inteins in newly sequenced genes (See Perler 1997 for a review). Criteria 2-4 help differentiate true inteins from in-frame inserts that result from inter-species sequence variability or other types of insertion sequences or protein editing. In the absence of experimentally demonstrating protein splicing, it should be emphasized that the combined use of these criteria, rather than the use of any single criterion, yields the most significant results.
1. An in-frame insertion in a gene that has a previously sequenced homolog lacking the insertion.
2. The observed size of the mature protein is similar to the size of homologs lacking the intein and not to the predicted size of the precursor. Many groups have gone a step further to prove protein splicing by amino acid sequencing across the splice junction in the ligated exteins or by identifying spliced peptides by mass spec analysis. In the absence of experimental proof of splicing, inteins should be considered putative and are marked theoretical in the Intein Registry.
3. The presence of intein splicing motifs consisting of Blocks A, N2, B, N4, F and G.
Although Blocks C, D, E and H are part of the endonuclease domain, they tend to be more conserved than the splicing motifs and are
sometimes easier to find in a candidate sequence. However, the presence of homing endonuclease domains is insufficient to classify a protein as an
intein, since many homing endonucleases are free-standing or found in introns. Mini-inteins that lack these DOD motifs are thus harder to
identify, especially when they contain non-consensus sequences in conserved positions. Note that recent papers have reported 'protein splicing' that is not intein-mediated, nor is it self-catalytic. Please distinguish between intein-mediated protein splicing and other Protein Editing mechanisms that result in spliced, rearranged proteins.
4. The presence of the four conserved splice junction residues:
- Ser, Thr or Cys
at the intein N-terminus
The dipeptide His-Asn or His-Gln at the intein C-terminus
Ser, Thr or Cys following the downstream splice site.
Ser, Thr, Cys and Asn are essential residues that act as nucleophiles in the splicing pathway.
The absence of these residues or the substitution with residues that cannot perform similar chemistry, would suggest an inactive
intein or an alternate splicing pathway. Thr has not been observed at the intein N-terminus, but can effectively substitute for Ser
in the Tli Pol-2 intein (Hodges 1992). The conserved Thr (Block B)
and His (in Blocks B and G) residues assist in catalysis and thus may not be essential since other residues in
the intein may provide similar facilitating functions in their absence (see Splicing mechanism).
Note that because of naturally occurring intein polymorphisms, not all active inteins contain all of
these conserved residues.
Intein polymorphisms:
1. Inteins have been identified with Ala at their N-termini
(see Splicing motifs) and splicing has been demonstrated in the KlbA family of Ala1 inteins (Southworth 2000) and the DnaB family of Ala1 inteins (Yamamoto 2001). These inteins splice by an second protein splicing mechanism (see Splicing mechanism) involving the direct attack on the peptide bond at the N-terminal splice junction by the Ser, Thr or Cys at the C-terminal splice junction (Xu 1994 and Southworth 2000).
2. Several inteins do not have a His as the penultimate residue (see Splicing motifs).
Some of these inteins have been shown to splice (Wu 1998, Chen 2000 and Scott 2000), while others did not splice in E.coli (Wang 1997). Except for the Ceu ClpP intein, all inteins tested that naturally lack a penultimate His are capable of splicing. However, some splice more efficiently than others. Splicing of the Mja PEP intein improved when its penultimate Phe was changed to His, but splicing of the Mja Rpol A' intein was inhibited when its penultimate Gly was changed to His (Chen 2000). We propose that inteins lacking a penultimate His (a) arose by mutation from ancestors in which a penultimate His facilitated splicing, (b) that loss of this His inhibited, but may not have blocked splicing and (c) that selective pressure for efficient expression of the RNA polymerase yielded an intein which utilizes another residue to assist Asn cyclization, changing the intein active site so that a penultimate His now inhibits splicing. This led to the hypothesis that the differences in splicing capacity of inteins that naturally lack a penultimate His may reflect inteins at different stages of evolving towards rapid splicing after mutation of their penultimate His and that splicing of inteins that naturally lack a penultimate His may improve if the native penultimate residue is replaced by His (Chen 2000).
3. A few inteins have been identified with a C-terminal Gln (Q)
(Pietrokovski 1998B,
Amitai 12004)
or a C-terminal Asp (D) (Amitai 12004),
(see Splicing motifs). Gln and Asp are capable of cyclization using similar mechanisms
as Asn, and should be able to substitute for Asn in the standard protein splicing pathway. However, several lines of evidence
suggest a modified protein splicing pathway for Gln and Asp inteins (Amitai 12004).
Return to Top
Intein Regions and Domains
- Three Regions are Found in Each Intein: an N-terminal Splicing Region
a central Homing Endonuclease Region or a small central Linker Region
a C-terminal Splicing Region.
-
Remarkably, inteins as small as 134 amino acids can splice out of precursor proteins
(Evans 1999). These 'mini-inteins' inteins do not have intein Blocks C, D, E, and H. The discovery of mini-inteins and mutational analysis have indicated that the residues responsible for protein splicing are present in the N-terminal Splicing Region and the C-terminal Splicing Region (including the +1 amino acid in the C-extein). The N-terminal Splicing Region is ~100 amino acids and begins at the intein N-terminus and ends shortly after Block B. The intein C-terminal Splicing Region is usually less than 50 amino acids and includes Blocks F and G.
The N-terminal Splicing Region and the C-terminal Splicing Region form a single structural domain, which is
conserved in all inteins studied to date (Duan 1997,
Klabunde 1998, Hall 1997<,
Poland 2000<,
Ichiyanagi 2000, reviewed in
Perler 1998.
Mini-inteins are usually ~130-200 amino acids. However, most inteins are greater than 300 amino acids, while the Pab RFC-2 intein is 608 amino acids (see Selected Intein Characteristics). These big inteins have a larger linker region between intein Blocks B and F that includes intein Blocks C, D, E, and H homing endonuclease motifs.
There are 4 classes of homing endonucleases that are defined by their conserved signature sequence motifs (Belfort 1997,
Mueller 1994, Jurica1999):
the DOD (also known as dodecapeptide or LAGLIDADG) family the HNH (or His-Asn-His) family the GIY-YIG family the His-Cys family
Besides the core endonuclease domain containing the DOD motifs, the Sce VMA intein also has a DNA recognition region (DRR) inserted before the DOD domain (Duan 1997,
but most other inteins do not appear to have a DRR at this position. The Pfu-RIR1-1 intein has a 'Stirrup' domain inserted after the DOD domain and before the C-terminal Splicing Domain (Ichiyanagi 2000).
A few inteins have HNH class homing endonucleases between intein Blocks B and F (See
Selected intein characteristics or LAGLIDADG (DOD) Homing Endonuclease Motifs).
Some inteins have large linker regions of unknown function, which may be the remnants of decaying homing endonucleases. It is interesting to note that 2 inteins (Cau Hyp intein and Ssp DnaX intein) have DOD homing endonuclease motifs present in a large linker region that is in a different reading frame than the remainder of the inteins. At this time, we do not know if these out of frame homing endonuclease genes are expressed either by a frame shifting mechanism or from an independent promoter. Alternatively, they may be homing endonuclease remnants that have undergone deletions and are never expressed in the correct reading frame for the homing endonuclease.
Homing endonuclease activity has only been tested in a small number of inteins and many of those tested are bifunctional proteins mediating both protein splicing and DNA cleavage (look for the 'PI-" name in the
Selected Intein CharacteristicsSection).
Return to Top
Intein Motif Consensus Sequences:
- The consensus sequence for each block is indicated below. Although, no single residue is invariant,
the Ser and Cys in Block A, the His in Block B, the His, Asn and Ser/Cys/Thr in Block G are the most conserved
residues in the splicing motifs. Any member of an amino acid group may be present in the
remaining positions, even when a specific predominant residue is indicated
(Pietrokovski 1994; Perler 1997
; http://blocks.fhcrc.org/~pietro/inteins;
Intein Motifs).
Remember, the splicing domain consists of Blocks A, N2, B, N3, F and G, while Blocks C, D, E and H are only in the
DOD homing endonuclease domains. Blocks C and E each contains an endonuclease active site Asp (D) or Glu (E).
Block D contains an endonuclease active site Lys (K) in the Sce VMA intein (Duan 1997).
Several inteins have mutations in these endonuclease active site residues and therefore may not be active endonucleases although
the remainder of the motif is present.
-
Key: upper case letters represent the standard single letter amino acid code for the most common amino acid at this position
and lower case letters represent
amino acid groups: ., any residue; h, hydrophobic residues: G,A,V,L,I, M; p, polar residues: S,C,T; a, acidic residues: D or E; r, aromatic residues: F,Y,W)
|
Block A: Ch..Dp.hhh..G |
(first residue = intein N-terminus) |
|
Block B : G..h.hT..H.hhh |
(usually 70-105 residues from N-terminus) |
|
Block C : LhG..hhaG |
(motif of DOD homing endo) |
|
Block D : .K.IP..h |
(motif of DOD homing endo) |
|
Block E : .L.GhFahDG |
(motif of DOD homing endo) |
|
Block H : p.S..hh..h..LL..hGI |
(motif of DOD homing endo) |
|
Block F : rVYDLpV[1-3 residues]a..[H or E]NFh |
|
|
Block G : NGhhhHNp |
(p = downstream extein N-terminus) |
Return to Top
|

