|
InBase Reference: Perler, F. B. (2002). InBase, the Intein Database. Nucleic Acids Res. 30, 383-384.
 This page has been added to serve as a single source page for basic intein information and figures. This page has been added to serve as a single source page for basic intein information and figures.
What is Protein Splicing?
Naming inteins
Intein Motifs
Protein Splicing and Homing Endonuclease Domains
Inteins as Mobile Genetic Elements: Homing Endonuclease Activity
Intein Alleles and the Phylogenetic Distribution of Inteins
Inteins are Related to Hedgehog Protein Autoprocessing domains
Evolution of Inteins and Hedgehog Proteins
Also see The Mechanism of Protein Splicing Page
Also see the Conserved Intein Features - Do you Have an Intein? Page
This section is still under construction. More figures will be added in the future.
See the Splicing
Mechanism page to download the Protein Splicing Mechanism animation.
Slide 1: Protein Splicing is Post-transcriptional
Slide 2: RNA vs. Protein Splicing
Slide 3: Intein Regions and Motifs: DOD intein vs. mini intein
Slide 4: DOD Intein Regions and Motifs
Slide 5: The Standard Protein Splicing Mechanism
Slide 6: The Standard and Alternate Protein Splicing Mechanisms
Slide 7: The Hedgehog Protein Autoprocessing Mechanism
Slide 8: Intein Evolution.
A. Protein Splicing Basics:
What is Protein Splicing?
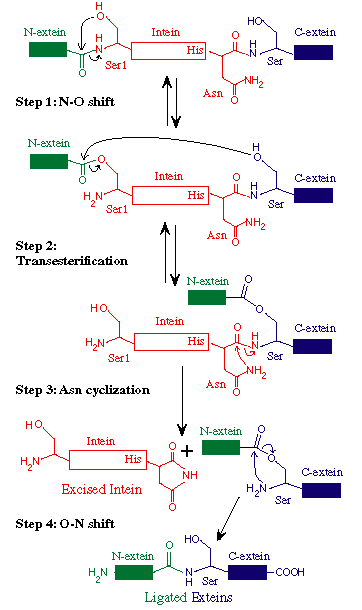
Protein splicing is defined as the excision of an intervening sequence (the INTEIN) from a protein precursor and the concomitant ligation of the flanking protein fragments (the EXTEINS) to form a mature host protein (extein) and the free intein (Perler 1994). Intein-mediated protein splicing results in a native peptide bond between the ligated exteins (Cooper 1993). Extein ligation differentiates protein splicing from other forms of autoproteolysis and conserved intein motifs differentiate inteins from other types of in-frame sequences present in one homolog and absent in another homolog.
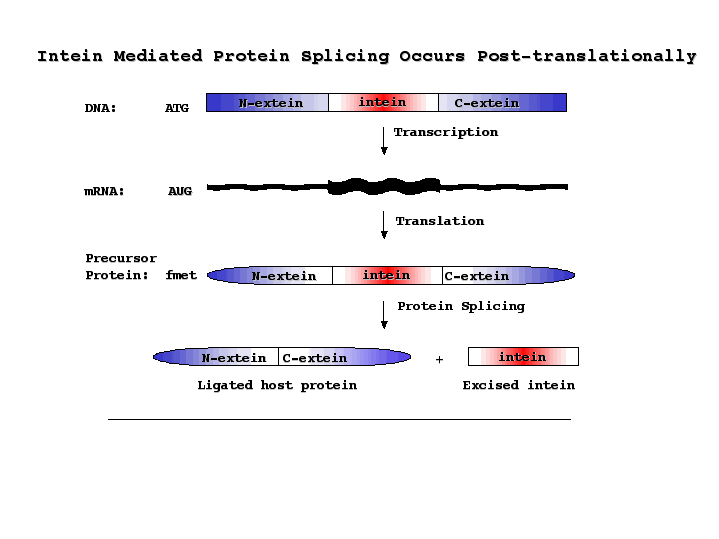
Download PDF Figure.
Return to Top
Inteins have sometimes been called 'Protein Introns'. Introns are intervening sequences that are spliced out of RNA before the mRNA is translated into a protein. The intron and the exon usually do not form a single open reading frame (ORF). During intein-mediated protein splicing, the intervening sequence is both present in the mature mRNA and translated to form a precursor protein. The intein is then spliced out of the precursor protein. The intein plus the first C-extein residue (called the +1 amino acid) contain sufficient information to mediate splicing of the intein out of the host protein and ligation of the exteins to form the active host protein. Many inteins can splice in heterologous foreign proteins if they are placed in a compatible host protein environment. The rules for what constitutes a good foreign extein environment are not well understood. It also appears that different inteins are more robust than others, with some inteins requiring many native extein residues and others only a single native C-extein amino acid.
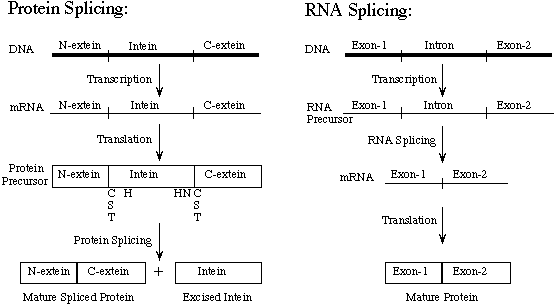
Download PDF Figure.
Naming inteins:
Inteins are named after the organism and gene in which they are found. The organism name follows the same consensus as restriction enzymes and uses a 3 letter genus + species designation, followed by a strain designation, if necessary. The organism name is followed by an abbreviation of the extein name. If more than 1 intein is present in an extein gene, the inteins are given a numerical suffix starting from 5' to 3' or in order of their identification.
For example, the Pyrococcus furiosus ribonucleoside-diphosphate reductase alpha subunit gene contains 2 inteins. The organism is abbreviated as 'Pfu'. Since the gene has been called the 'RIR1' gene, the inteins are named using this gene name. Thus, these 2 inteins are called the Pfu RIR1-1 intein inserted after Gly 301 in the Pfu RIR1 precursor protein and the Pfu RIR1-2 intein inserted after Pro914 in the Pfu RIR1 precursor protein.
Note that an intein name, such as the Pfu RIR1-1 intein, refers to both the intein gene and the intein protein. In many publications, the consensus is to italicize the gene name and to capitalize the first letter of the protein name.
As described below, some inteins are bifunctional proteins that also have endonuclease activity. When endonuclease activity has been demonstrated, the intein is also given a second name that follows the endonuclease naming conventions (Belfort 1997). This name includes the prefix 'PI-', the 3 letter organism abbreviation and a Roman numeral indicating the order of identification of the intein endonuclease in that organism. The endonuclease names for the Pfu RIR1- and Pfu RIR1-2 inteins are PI-PfuI and PI-PfuII, respectively.
There is also a convention for numbering amino acids in inteins. Although we often number the residues in the precursor as a single protein, as when intein insertion site locations are given, a second numbering scheme is often used to assist thinking about inteins in heterologous or foreign exteins. The intein amino acids are numbered from N-terminal to C-terminal beginning with the first residue of the intein and ending with the last residue of the intein. The amino acids in the N-extein: (a) start with the number 1, (b) include a minus sign prefix and (c) are counted from right to left (beginning with the last N-extein residue and going towards the N-terminus). The amino acid preceding the intein is the -1 amino acid. The amino acids in the C-extein: (a) are numbered beginning at the C-terminal splice junction, (b) include a plus sign prefix and (c) are counted from amino to C-terminus.
The first residue following the intein is the mechanistically essential +1 amino acid, which is not technically part of the intein since the intein is defined as the intervening sequence that is spliced out of the precursor.
Return to Top
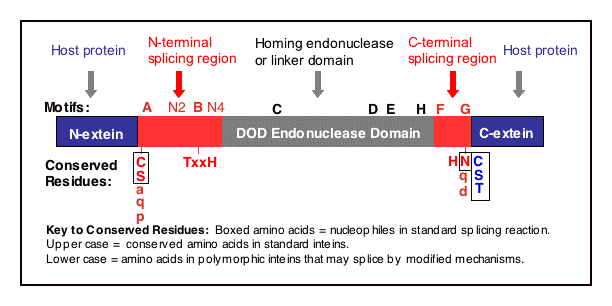
Intein Motifs:
Several conserved motifs have been observed by comparing intein amino acid sequences. There are two nomenclatures for these motifs: Blocks A, B, C, D, E, H, F, G (Pietrokovski 1994 and Perler 1997) or Blocks N1, N3, EN1, EN2, EN3, EN4, C2 and C1, respectively (Pietrokovski 1998). Blocks N2 and N4 are not as well conserved as the other intein motifs (Pietrokovski 1998). The intein motifs are more extensively described in the Conserved Intein Features - Do you Have an Intein? section and their sequences are listed in the Splicing Motifs
and LAGLIDADG (DOD) Homing Endonuclease Motifs sections.
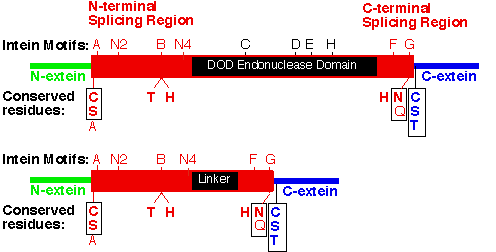
Download PDF Figure
Figure Legend. Intein regions and conserved motifs.
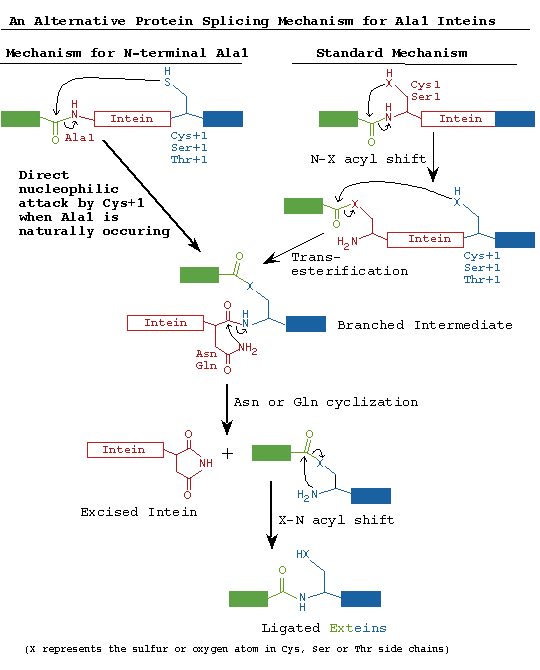
An intein with a DOD homing endonuclease is depicted with conserved motifs listed above and conserved residues involved in catalysis shown below. Nucleophiles are boxed. Many of the most conserved residues participate in the protein splicing reaction, which is described in the Mechanism of Protein Splicing Section. The conserved Ser, Thr or Cys on the C-terminal side of both splice junctions can mediate similar chemical reactions. Likewise, Asn and Gln can perform similar cyclization reactions. The Thr and His in Block B assist reactions at the N-terminal splice junction and the His in Block G assists reactions at the C-terminal splice junction. The splicing regions (red boxes) are separated by a linker or a homing endonuclease region. Polymorphism in nucleophiles and assisting groups is becoming increasingly more evident as more inteins have been sequenced. A second protein splicing mechanism has been described for inteins that naturally contain an N-terminal Ala (Southworth 2000).
Return to Top
Protein Splicing and Homing Endonuclease Domains
Three Regions are Found in Each Intein:
an N-terminal Splicing Region
a central Homing Endonuclease Region or a small central Linker Region
a C-terminal Splicing Region.
Remarkably, inteins as small as 134 amino acids can splice out of precursor proteins (Evans 1999). These 'mini-inteins' inteins do not have intein Blocks C, D, E, and H. The discovery of mini-inteins and mutational analysis have indicated that the residues responsible for protein splicing are present in the N-terminal Splicing Region and the C-terminal Splicing Region (including the +1 amino acid in the C-extein). The N-terminal Splicing Region is ~100 amino acids and begins at the intein N-terminus and ends shortly after Block B. The intein C-terminal Splicing Region is usually less than 50 amino acids and includes Blocks F and G.
The N-terminal Splicing Region and the C-terminal Splicing Region form a single structural domain, which is conserved in all inteins studied to date (Duan 1997, Klabunde 1998, Hall 1997, Poland 2000, Ichiyanagi 2000, reviewed in Perler 1998).
Most inteins are greater than 300 amino acids, while the Pab RFC-2 intein is 608 amino acids (see Selected Intein Characteristics). These big inteins have a larger linker region between intein Blocks B and F. Almost all of these inteins include intein Blocks C, D, E, and H, which are shared with the DOD family of homing endonucleases.
There are 4 classes of homing endonucleases that are defined by their conserved signature sequence motifs (Belfort 1997, Mueller 1994, Jurica1999):
the DOD (also known as dodecapeptide or LAGLIDADG) family
the HNH (or His-Asn-His) family
the GIY-YIG family
the His-Cys family
Besides the core endonuclease domain containing the LAGLIDADG motifs, the Sce VMA intein also has a DNA recognition region (DRR) inserted before the DOD domain (Duan 1997), but most other inteins do not appear to have a DRR at this position. The Pfu-RIR1-1 intein has a 'Stirrup' domain inserted after the DOD domain and before the C-terminal Splicing Domain (Ichiyanagi 2000).
A few inteins have HNH class homing endonucleases between intein Blocks B and F (See Selected intein characteristics or LAGLIDADG (DOD) Homing Endonuclease Motifs). Some inteins have large linker regions of unknown function, which may be the remnants of decaying homing endonucleases. It is interesting to note that 2 inteins (Cau Hyp intein and Ssp DnaX intein) have DOD homing endonuclease motifs present in a large linker region that is in a different reading frame than the remainder of the inteins. At this time, we do not know if these out of frame homing endonuclease genes are expressed either by a frame shifting mechanism or from an independent promoter. Alternatively, they may be homing endonuclease remnants that have undergone deletions and are never expressed in the correct reading frame for the homing endonuclease.
Homing endonuclease activity has only been tested in a small number of inteins and many of those tested are bifunctional proteins mediating both protein splicing and DNA cleavage (look for the 'PI-" name in the Selected Intein Characteristics Section).
Return to Top
Inteins as Mobile Genetic Elements: Homing Endonuclease Activity
Homing endonucleases were first studied as part of mobile introns. Homing endonucleases make double-strand breaks in DNA at or near the insertion site (home) of intein or intron genes in host protein alleles that lack the intein or intron (Belfort 1997, Mueller 1994, and Jurica 1999).
Homing endonuclease activity initiates intein gene mobility into intein-less extein alleles (Lambowitz 1993 and Belfort 1995, Gimble 2000 and Jurica1999). If an intein-less allele of the extein gene enters the cell as the result of sex, conjugation, infection, transformation or any other means, the intein gene can mobilize into the intein-less extein gene. This gene conversion event involves double-strand break repair mechanisms and is initiated by the endonuclease activity of the intein. Once the homing endonuclease cleaves the intein-less extein gene, the only copy of the gene remaining for repair of the DNA break is the intein-containing gene. Thus, gene conversion from intein-minus to intein-plus is very efficient when the intein is also a functional homing endonuclease (Gimble 1992).
The homing endonuclease recognition site is usually very large compared to regular restriction enzymes, often 20-40 nucleotides. The site is only present once in the genome of organisms which lack the intein (Belfort 1997 and Jurica 1999). The homing endonuclease recognition site is not present in the genome of an organism that contains the intein, since the intein coding region interrupts the homing endonuclease recognition site.
Return to Top
Intein Alleles and the Phylogenetic Distribution of Inteins
The phylogenetic distribution of inteins is sporadic. The presence of an intein in a particular gene does not necessarily mean that an extein homolog from a closely related species or strain will have the same intein. For example, look at the inteins present in the 3 insertion sites in DNA polymerases from various strains of archaea or GyrA inteins in various species of Mycobacterium. At this time, it is not clear whether this pattern of intein distribution represents loss of ancient inteins or more recent acquisition of inteins due to intein gene mobility. In many cases analyzed, the codon usage and GC content of the intein coding region is different from the surrounding extein coding region, suggesting recent horizontal transmission. Organisms that have a large number of inteins may have acquired this large number of inteins because they (1) can easily take up DNA from the environment (naturally competent), (2) share viruses, conjugative elements, plasmids, etc. that have broad host ranges, or (3) have very efficient gene conversion machinery, double-stand breaks repair systems and/or recombination systems.
Several inteins, such as the DnaB, RIR1, GyrA and Pol inteins, are present at the same extein insertion site in extein homologs from several species, including extein homologs in organisms from different phylogenetic domains. Perler 1997 suggested that inteins present in the same insertion site of an extein homolog be considered intein alleles or homologs.
Inteins grouped by extein insertion site are tabulated in the Intein Alleles Section and extein insertion sites are named according to
http://bioinformatics.weizmann.ac.il/~pietro. Intein alleles are more closely related to each other than to other inteins in the same organism or even in the same gene.
Extein proteins may also have multiple inteins present at different insertion sites within the extein (for example, Tli Pol, Tsp-TY Pol, Mja RFC, Mja RNR or Pfu RIR1). A few proteins have 3 inteins.
Intein alleles are more closely related to each other than to other inteins because they are either descendants of an ancestral intein or have been recently mobilized into that site based on homing endonuclease specificity. Another way of saying this, is that inteins that have the same homing endonuclease specificity are more related to each other than to other inteins. Remember, the homing endonuclease recognition site determines the intein insertion site because the double-strand break made by the homing endonuclease initiates the site-specific gene conversion reaction leading to intein acquisition.
Inteins are Related to Hedgehog Protein Autoprocessing domains
Crystal structures have been determined for several inteins:
Sce VMA intein (Duan 1997)
Mxe GyrA intein (Klabunde 1998)
Pfu RIR1-1 intein (Ichiyanagi 2000).
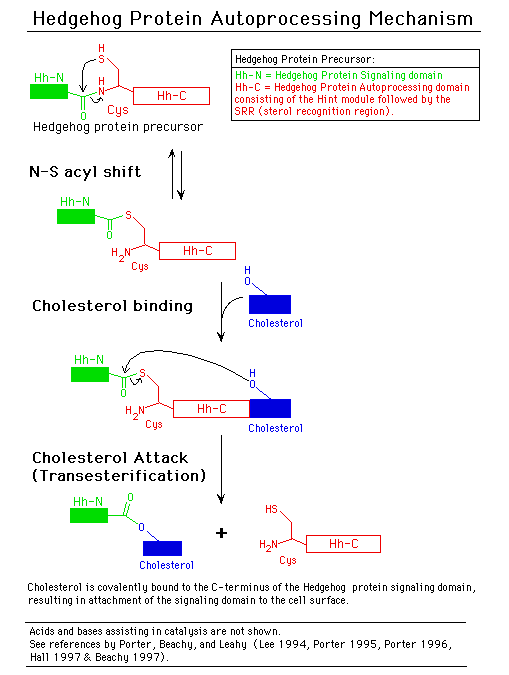
The main chain trace of the splicing region of these inteins conforms to the beta-strand structure of the Drosophila hedgehog protein autoprocessing domain (Hall 1997), although the 4 proteins have little amino acid sequence similarity. This led Leahy and coworkers to call this new protein fold the HINT module (Hedgehog and INTein) (Hall 1997). The hedgehog proteins direct embryonic pattern formation in numerous multicellular organisms (Beachy 1997). Several genes in Caenorhabditis elegans genes have domains similar to HINT modules, but with different N-terminal functional domains (Aspock 1999).
The similarity between inteins and hedgehog protein autoprocessing domains goes beyond structure to include common conserved sequence motifs and biochemical functions (Koonin 1995, Beachy 1997, Dalgaard 1997, Perler 1997C, Perler 1998 and Pietrokovski 1998). Both types of proteins mediate autoprocessing reactions initiated by an acyl shift to form an activated (thio)ester bond that is cleaved via transesterification by different thiol or hydroxyl containing molecules (see the Mechanism of Protein Splicing section, and Beachy 1997 and Perler 1997C). Thus, higher organisms have redirected the ability of inteins to ligate flanking peptides and utilized modified inteins to ligate lipids to the hedgehog signaling domain for compartmentalization at the cell surface (which is required for signaling).
Return to Top
Evolution of Inteins and Hedgehog Proteins
The function of the progenitor HINT module may have been the formation of a reactive (thio)ester bond at the C-terminus of the fused target polypeptide. This linkage could then be directly attacked by numerous types of nucleophiles present in polypeptides or other molecules, resulting in ligation of the attacking moiety to the C-terminus of the target polypeptide and release from the HINT module. Initially, this ligation event may have occurred in trans with randomly associating molecules and could have been an early means of generating larger proteins prior to the development of sophisticated recombination systems or of adding post-translational modifications. Subsequently, residues C-terminal to the core HINT module were added for selection and alignment of the attacking molecule to the target polypeptide. The Sterol Recognition Region (SRR) at the C-terminus of the Hedgehog HINT module is required for ligation of cholesterol to the Hedgehog signaling domain, while the Peptide Ligation Region at the C-terminus of the intein HINT module is required for ligation of a fused polypeptide (C-extein) to the target polypeptide (N-extein). Caenorhabditis elegans has several genes with HINT domains linked to various SRR modules and N-terminal domains of unknown function (Hall 1997 and Aspock 1999). It is also possible that PLR and SRR regions evolved in steps, with residues first being added C-terminal to the core HINT module followed by further evolution into functional PLR and SRR regions. Finally, it is likely that inteins have both acquired and lost endonuclease domains during evolution. Insertion of mobile homing endonuclease genes into intein genes would afford the endonuclease a safe refuge, since splicing would preserve host gene function.
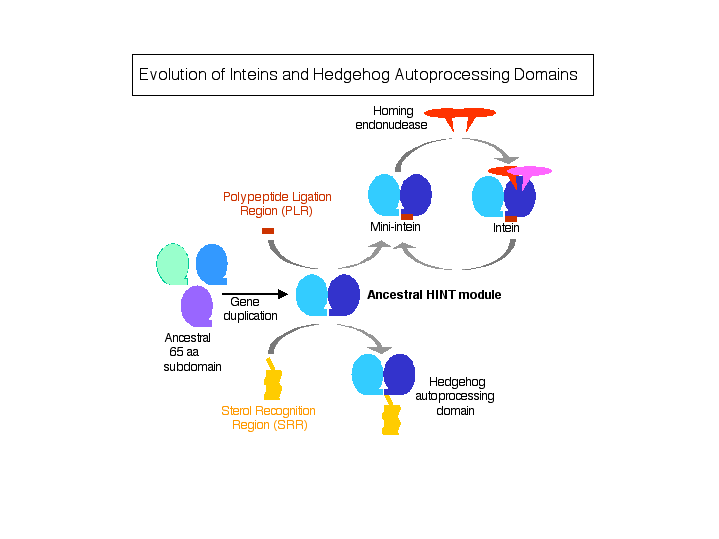
Download PDF Figure.
Figure Legend. Evolution of inteins and Hedgehog-like autoprocessing proteins. Schematic drawing illustrating the events that may have occurred during evolution of Hedgehog protein autoprocessing domains and inteins. Prior to duplication, the 65 amino acid (aa) module may have functioned as a dimer. For clarity, polypeptides at the N-terminus of the HINT module and molecules to be ligated to these target polypeptides are not depicted.
Return to Top
B. Teacher's packet - download intein slides
Note: this section is still under construction.
Slide 1: Protein Splicing is Post-transcriptional

Download Splicing PDF
Slide 2: RNA vs. Protein Splicing

Download RNA vs. Protein Splicing PDF
Slide 3: Intein Regions and Motifs: DOD intein vs. mini intein

Download Mini-intein and DOD Intein PDF
Slide 4: DOD Intein Regions and Motifs
Download DOD Intein PDF
Slide 5: The Standard Protein Splicing Mechanism
Download Mechanism PDF
Slide 6: The Standard and Alternate Protein Splicing Mechanisms
Download Alternate Mechanism PDF
Slide 7: The Hedgehog Protein Autoprocessing Mechanism
Download Hedgehog Mechanism PDF
Slide 8: Intein Evolution.

Download Intein Evolution PDF
|

