F. Mechanism References
The mechanism of intein-mediated protein splicing
Protein splicing is so rapid that the precursor protein is rarely observed
in native systems. The intein plus the first C-extein residue contain sufficient
information for splicing in foreign proteins. However, exteins may affect
splicing rates or efficiency. Splicing in foreign protein contexts often
results in an increase in dead-end cleavage reaction products.
Protein splicing involves 4 nucleophilic displacements
by the 3 conserved splice junction residues.
Acids and bases or hydrogen bonding residues that assist these nucleophilic
displacements are omitted in the figure below. The intein
penultimate His in Block G assists in Asn cyclization and C-terminal
cleavage (Xu
1996) by hydrogen bonding to the Asn carbonyl oxygen, making this peptide
bond more labile (Klabunde
1998, Duan
1997). The Thr and His in Block B assist
in the initial acyl rearrangement at the N-terminal splice junction (Kawasaki
1997) by hydrogen bonding to main chain atoms and holding the residue
preceding the intein in a non-standard cis conformation (Klabunde
1998) or in a strained conformation (Poland
2000). Any residue that can form similar hydrogen
bonds can substitute for these conserved facilitating residues in Blocks
B and G. The mechanism of protein splicing has recently been reviewed
in Noren
2000, Paulus
2000, Perler
1997C, Shao
1997 and Perler
1998. Several previous reviews contain mechanisms now known to be incorrect.
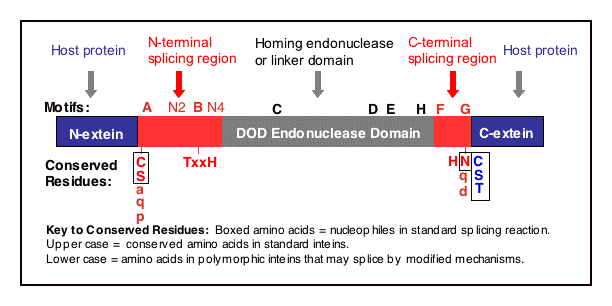
A. The protein splicing mechanism depicted with Ser at
both splice junctions
STEP 1: The N-terminal splice junction is activated by a N-O
or N-S acyl rearrangement at the intein N-terminus that moves the
N-extein to the side chain of the Ser/Cys at the intein N-terminus, forming
the linear ester/thioester intermediate. A few inteins have been identified
with a N-terminal Ala (A) (see Splicing
motifs), although splicing has not been demonstrated with these inteins.
Ala cannot undergo an acyl shift like Ser/Thr/Cys, since it doesn't have
an hydroxyl/thiol side chain. However, these inteins may be active if the
residues facilitating the reaction are still making the splice junction
peptide bond more labile and if the C-extein Ser/Thr/Cys is in the proper
position to attack the splice site; in this case, the downstream splice
junction Ser/Thr/Cys would directly cleave the N-terminal splice junction
peptide bond (see splicing pathway A in Xu
1994) to form the branch intermediate.
STEP 2: The upstream ester/thioester bond is attacked during
a transesterification reaction by the hydroxyl/thiol
group of the C-extein Ser/Thr/Cys, resulting in cleavage at the N-terminal
splice junction and transfer of the N-extein to the side chain of the C-extein
Ser/Thr/Cys, forming the branched protein intermediate.
STEP 3: The branch is resolved by cyclization
of the conserved intein C-terminal Asn to form a succinimide ring,
resulting in cleavage of the C-terminal splice junction. The succinimide
can be hydrolyzed to form Asn or isoasparagine. A few inteins have been
identified with a C-terminal Gln (Q) (see
Splicing motifs);
although splicing has not been demonstrated with these inteins, Gln is
capable of undergoing a cyclization reaction just like Asn and should thus
be able to substitute for Asn.
STEP 4: A spontaneous 0-N or S-N acyl rearrangement
results in formation of a native peptide bond between the exteins.
Return to Top
B.  Animation of The Protein Splicing Pathway
Animation of The Protein Splicing Pathway
1. See the animation of The Protein Splicing Pathway with FLASH
or QuickTime.
2. Click here to download the Animation of The Protein Splicing Pathway.
Please Note: the PowerPoint 98 (Macintosh) animation of the protein splicing mechanism will automatically open on some browsers. However, with other browsers you may have to manually start the PowerPoint slide show (as you normally would any PowerPoint presentation) or first download the file and then run it in PowerPoint.
Return to Top
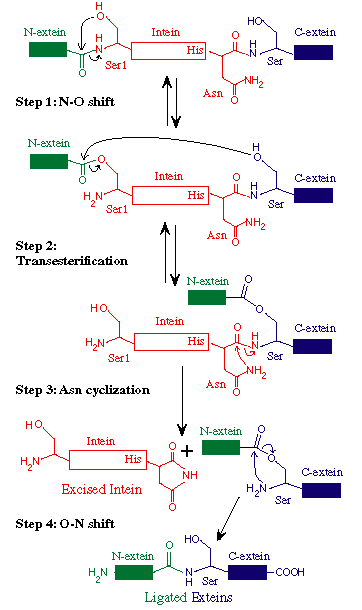
C. An Alternative Protein Splicing Mechanism for Inteins
that Naturally Begin with Ala.
Variations in the intein-mediated protein splicing mechanism are becoming
more apparent as polymorphisms in conserved catalytic residues are identified.
Several families of inteins have been identified
that begin with Ala rather than the consensus nucleophiles, Ser or Cys.
In standard inteins, an N-terminal Ser, Cys or Thr is absolutely required
for splicing. An N-terminal Ala cannot perform the initial reaction of
the standard protein splicing pathway to yield the requisite N-terminal
splice junction (thio)ester. However, experiments with the M. jannaschii
KlbA intein demonstrated that Ala1 inteins can splice efficiently using
an alternative protein splicing mechanism (Southworth
2000). In this non-canonical pathway, the C-extein nucleophile (Ser,
Cys or Thr) attacks a peptide bond at the N-terminal splice junction rather
than a (thio)ester bond, alleviating the need to form the initial (thio)ester
at the N-terminal splice junction. The remainder of the two pathways is
identical: branch resolution by Asn cyclization is followed by an acyl
rearrangement to form a native peptide bond between the ligated exteins.
Just like standard inteins, the Mja KlbA intein also requires the help
of the conserved Thr and His in Block B to activate the N-terminal splice
junction. We have also demonstrated splicing of the Mle DnaB intein (dnaB-b
insertion site, E. Davis, M. Southworth & F. Perler, unpublished data)
which is another Ala1 intein, suggesting that different families of naturally
occurring Ala1 inteins should be capable of splicing.
The KlbA and Mle DnaB inteins have overcome the barriers to direct nucleophilic
attack on the peptide bond at the N-terminal splice junction that are present
in previously studied inteins with Ser or Cys at their N-terminus. It is
unclear why other inteins can't perform similar reactions, since the Block
B oxyanion hole is still available to facilitate direct attack on the N-terminal
splice junction. Possibly, (thio)ester formation may be necessary in standard
inteins to align the C-extein nucleophile, to remove steric hindrances
or to induce a conformational shift that allows attack by the +1 nucleophile
(Cys, Ser or Thr). The crystal structure of a S.cerevisiae VMA intein precursor
has helped to resolve this question by revealing that Cys+1 is too far
away to directly attack either a peptide or a thioester bond at the N-terminal
splice junction, leading the authors to suggest that inteins must undergo
a conformational shift to allow attack by the Cys+1 nucleophile (Poland
2000). We propose that Cys+1 (or its equivalent residue) in Ala1 inteins
is already in position to attack the N-terminal splice junction amide bond
in the precursors protein.
Return to Top
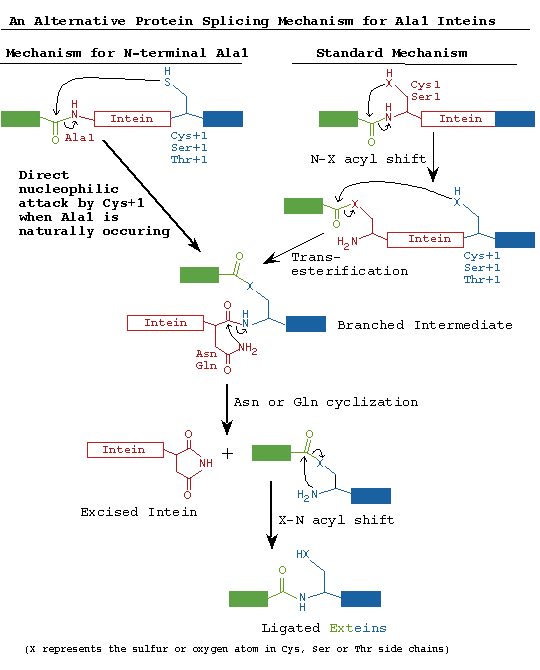
D. Similarities between inteins and hedgehog protein autoprocessing
domains
Hedgehog proteins are signaling molecules required for embryonic pattern
formation (Beachy
1997). They are synthesized as inactive precursors with an N-terminal
signaling domain linked to a C-terminal autoprocessing domain (Hh-C). Hh-C
begins with a Cys that undergoes an acyl rearrangement analogous to Step
1 of the protein splicing pathway. Hh-C also has sequence similarity to
inteins with conserved sequences corresponding to intein Blocks A and B
(Koonin
1995). In a transesterification reaction similar to Step 2 of the protein
splicing pathway, the hydroxyl group of cholesterol attacks this thioester
bond, resulting in attachment of cholesterol to the C-terminus of the hedgehog
protein signaling domain. Cholesterol anchors the signaling domain to the
cell surface. The Drosophila Hh-C domain is composed of a subdomain that
directs thioester formation, followed by a sterol recognition region required
for cholesterol transfer. Several nematode Hh-C domains contain unrelated
C-terminal extensions that may interact with molecules other than cholesterol
and have been tentatively termed Adduct Recognition Regions (Beachy
1997). Crystal structure analysis (see below) indicates that Hedgehog
autoprocessing domains evolved from a common ancestor and that higher organisms
redirected the ability of inteins to ligate flanking peptides and utilized
these modified inteins to ligate lipids to the hedgehog signaling domain
for compartmentalization at the cell surface (which is required for signaling).
Not only is there sequence and mechanistic similarities between inteins
and hedgehog protein autoprocessing domains, but there is a high degree
of structural identity amongst main chain alpha carbon atoms. This structural
similarity led Leahy and coworkers to propose that inteins and Hh-C have
a common structural fold (Hall
1997). This conserved structure is called a Hint
module (Hedgehog, INTein). The main chain alpha carbon
atoms of 100 amino acids in the Mxe GyrA intein are superimposable onto
the Hint module fold despite the fact that there is little amino acid sequence
identity (Klabunde
1998). Furthermore, Leahy and coworkers have proposed that inteins
and Hh-C evolved from a common precursor (Hall
1997 and Beachy
1997).
Hint modules are composed of ~12 beta-strands. In inteins, the core
endonuclease or linker region is inserted into the Hint module between
intein Blocks N4 and F. The Sce VMA intein has an additional endonuclease
DNA recognition region (DRR) between Blocks B and N4 (Duan
1997, Hall
1997 and Perler
1998) that is not present at this position in most other inteins. The
core endonuclease domain is composed of both beta-strands and alpha-helices.
The structure of the Sce VMA intein core endonuclease domain (Duan
1997) is very similar to the structure of a dimer of the intron encoded
endonuclease, I-CreI (Heath
1997). Both PI-SceI and I-CreI are members of the LAGLIDADG (DOD) family
of homing endonucleases.
-
Return to Top
F. Selected Mechanism References:
-
Hodges
1992
-
Xu 1993
-
Xu 1994
-
Shao 1995
-
Xu 1996
-
Chong 1996
-
Shao 1996
-
Duan 1997
-
Hall 1997
-
Heath 1997
-
Kawasaki
1997
-
Nogami
1997
-
Wang 1997
-
Shao 1997B
-
Anraku
1997B
-
Derbyshire
1997
-
Klabunde
1998
-
Perler
1998
-
Paulus1998B
-
Chong 1998
-
Wood 1999
-
Chen 2000
-
Noren 2000
-
Paulus
2000
-
Poland
2000
-
Southworth
2000
Return to Top

