|
InBase Reference: Perler, F. B. (2002). InBase, the Intein Database. Nucleic Acids Res. 30, 383-384.
The Endonuclease Motifs Page will not be updated after July 7, 2003
Please note: Finding DOD homing endonuclease motifs is not sufficient evidence that the gene contains an intein since DOD homing endonucleases are also present in introns and as free standing genes.
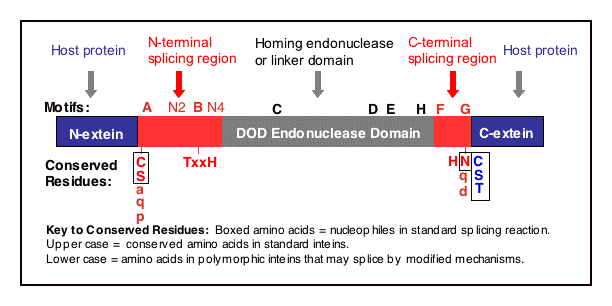
DOD (LAGLIDADG) Homing Endonuclease Motifs
The entire intein sequence is present in the individual intein files.
Central intein motifs C, D, E & H are in the DOD family core homing endonuclease domain (Duan 1997, Hall 1997, Klabunde 1998, Perler 1998, Perler 1997, Pietrokovski 1998, Dalgaard 1997, Belfort 1997, Jurica 1999 and Mueller 1994). These motifs form 4 conserved helices (Duan 1997 and Heath 1997). Blocks C and E are the original LAGLIDADG motifs and each contains an endonuclease active site Asp (D) or Glu (E). Block D contains a putative active site Lys (K), as observed in the Sce VMA intein (Duan 1997). Several inteins have mutations in these active site residues and therefore may not be active endonucleases although the remainder of the motif is present.
Mini-inteins are indicated as 'none' in Blocks C, D, E & H.
Dashes indicate that the individual motif ihas not been found.
The Ssp GyrB intein has an HNH family homing endonuclease between intein Blocks B and F.
The Ssp DnaX and Cau RIR1 inteins have a DOD homing endonuclease present in a different reading frame than the intein.
The position of the last amino acid in each block is listed to the right of the block.
An individual or amino acid group designation (see The Consensus Key below) in the consensus line indicates that the amino acid (upper case letter) or group (lower case letter) is present in a majority of the first 38 inteins sequenced, excluding the highly similar allelic inteins Perler 1997.
Dots in the consensus motifs indicate the position of non-conserved amino acids.
Please note: the absence of motif annotation indicates that the record was submitted without this information and it has yet to be added by the curator.
CONSENSUS LINE KEY:
| h | hydrophobic residues (G,V,L,I,A,M) |
| a | acidic residues (D,E) |
| r | aromatic residues (F,Y,W) |
| p | polar residues (S,T,C) |
| / | to align block, 1 or more AA not shown |
| - | motif absent |
| . | non-conserved residue |
| * | gap introduced into Block F |
| underlined residues | conserved in almost all inteins |
| capital letters | single letter amino acid code |
|

